UV Carcinogenesis
How do cells respond to UV damage? How does caffeine prevent skin cancer?
High burden of skin cancer in the U.S.
Skin cancer is the most prevalent cancer in humans and is associated with ultraviolet (UV). Annual incidence of skin cancer is 5.5 million in the U.S., and it exceeds all other cancers combined. Thus, there is a pressing need to identify and develop novel approaches to target UV-induced skin cancers.
UV induces DNA damage responses in cells
Ultraviolet (UV) from one-hour sunlight generates a surprisingly high number of lesions on DNA: 100,000 DNA lesions per cell that are potentially mutagenic, leading to the most prevalent cancers in humans. Understanding how cells respond to UV-induced DNA lesions could be helpful to selectively kill DNA-damaged cells and prevent UV-associated skin cancers.
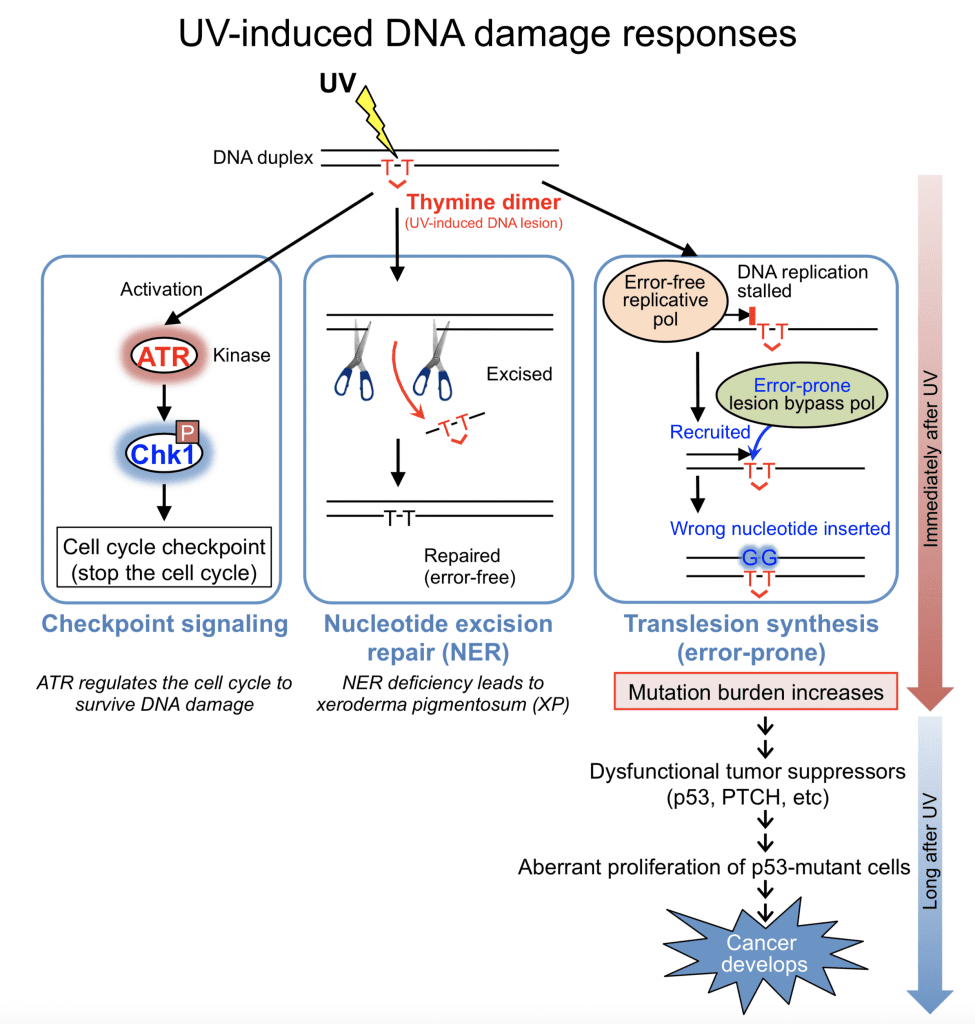
UV generates DNA lesions by forming dimers at dipyrimidine sites (two adjacent cytosine or thymine). There are two major types of UV-induced DNA lesions: cyclobutane pyrimidine dimers (CPD) and 6-4 photoproducts (6-4PP). These lesions are structurally distinct and activate at least three different biological processes: checkpoint signaling, nucleotide excision repair (NER), and translesion synthesis.
UV activates the ATR kinase that phosphorylates downstream targets including Chk1 to stop the cell cycle. This allows time for DNA repair. NER is an error-free repair mechanism for UV-induced DNA lesions. If DNA lesions are unrepaired, these DNA-distorting lesions stall DNA replication in S phase of the cell cycle because replicative DNA polymerases cannot synthesize DNA across the lesions. Specialized DNA polymerases are recruited to DNA-damaged sites in order to continue DNA synthesis across the lesions (translesion synthesis), but the lesion-bypass polymerases are error-prone. Wrong nucleotides are often inserted opposite the lesions, resulting in mutation incorporation. Chronic UV irradiation leads to accumulation of genetic mutations and skin cancer development. We aim to elucidate regulatory mechanisms underlying error-prone translesion synthesis.
Replication stress activates ATR
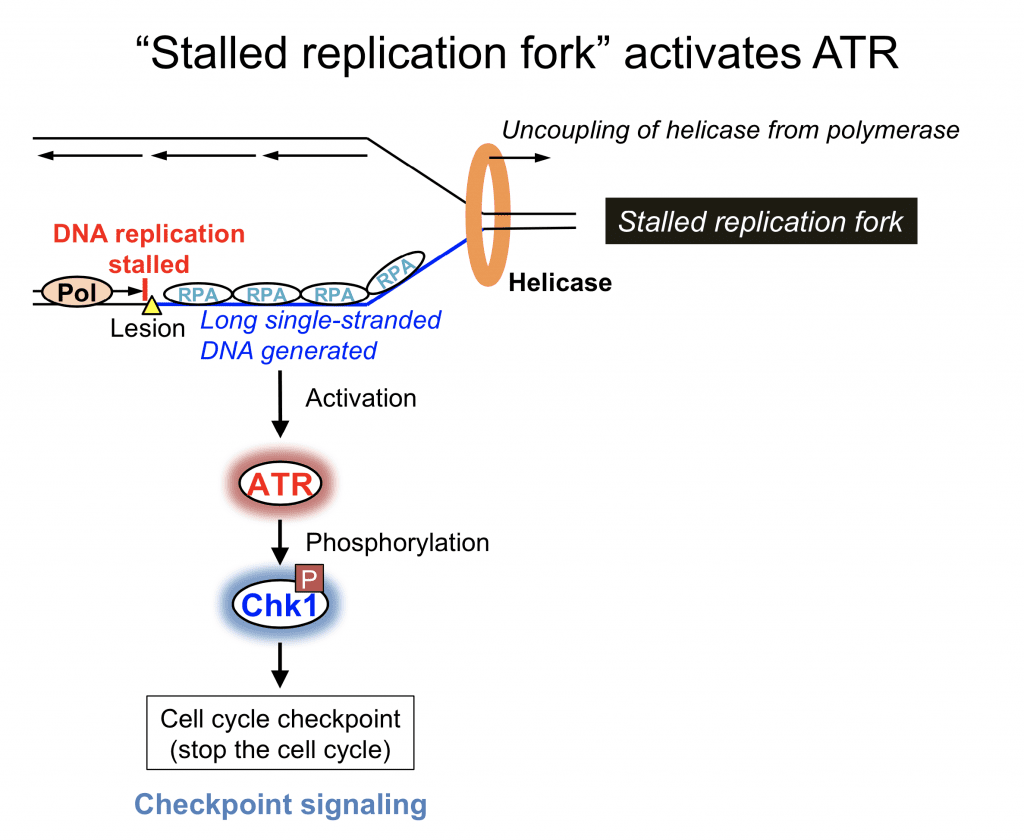
UV-induced DNA lesions cause replication stress because these lesions distort DNA and replicative DNA polymerases cannot continue DNA synthesis across the lesions. When DNA replication is stalled during S phase, a long stretch of single-stranded DNA is generated because DNA helicase continues unwinding DNA duplex (uncoupling of helicase from polymerase). This is a key structure to activate the ATR kinase. Many mediator proteins are recruited to DNA-damaged sites in order to form the ATR activation complex for full activation of the ATR kinase.
To better understand UV-induced DNA damage response, we recently dissected differential roles of two major types of UV-induced DNA lesions (CPD and 6-4PP) in ATR pathway activation and DNA replication blockage. This study provides insight into potential mechanisms by which caffeine (nonspecific ATR inhibitor) inhibits mutation incorporation following UV irradiation.
Caffeine prevents skin cancer
Sunscreen and sun avoidance are very effective and important to lower the risk of skin cancer. Also, dietary compounds that can prevent skin cancer are of great interest (chemoprevention).
Strikingly, multiple human epidemiological studies demonstrated that caffeinated coffee intake is associated with decreased risks of developing nonmelanoma skin cancer and melanoma. Each daily cup of caffeinated coffee is associated with 5% reduced risk of skin cancer. The cancer-preventive effect of caffeinated coffee is dose-dependent. People who drink 6 or more cups of caffeinated coffee per day showed ~30% reduced risk of skin cancer. Importantly, decaffeinated coffee has no such effects, indicating that caffeine is an active constituent for skin cancer prevention.
In parallel to human studies, the effects of caffeine on UV carcinogenesis have been extensively studied in mouse models. Both topical application and oral administration of caffeine suppress skin carcinogenesis induced by chronic UV irradiation.
References
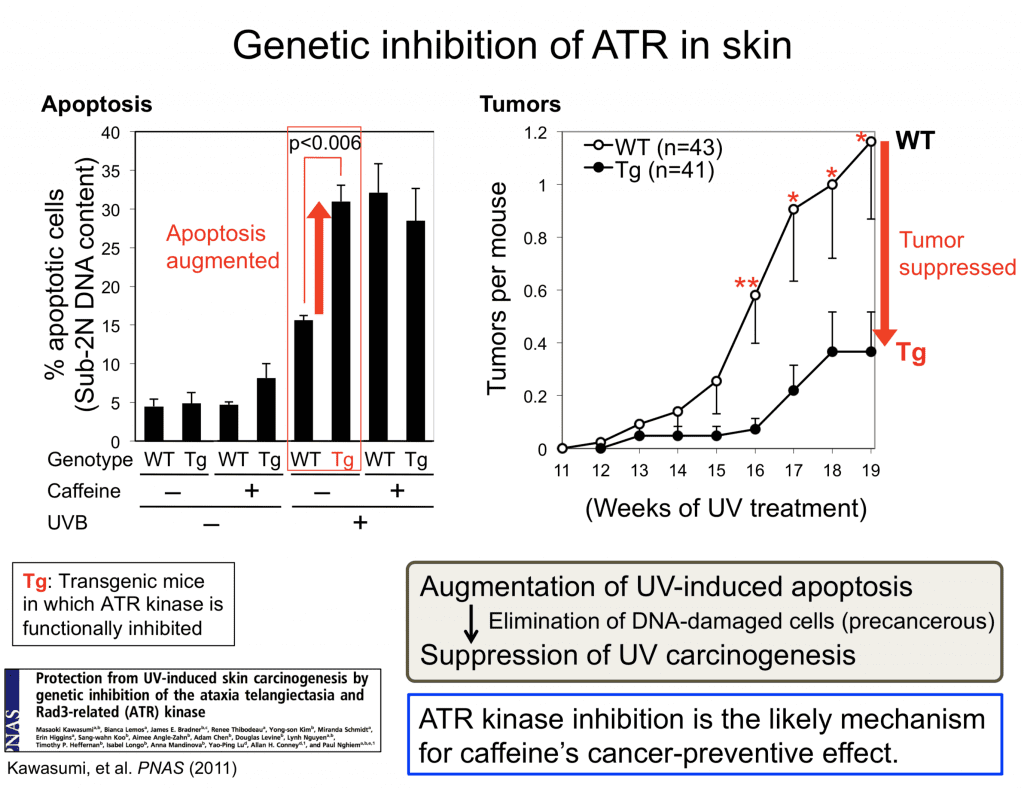
Protection from UV-induced skin carcinogenesis by genetic inhibition of the ataxia telangiectasia and Rad3-related (ATR) kinase
Kawasumi M, Lemos B, Bradner JE, Thibodeau R, Kim YS, Schmidt M, Higgins E, Koo SW, Angle-Zahn A, Chen A, Levine D, Nguyen L, Heffernan TP, Longo I, Mandinova A, Lu YP, Conney AH, Nghiem P
Proc Natl Acad Sci U S A 2011;108(33):13716-13721
PMID: 21844338
Mechanisms of caffeine-induced inhibition of UVB carcinogenesis
Conney AH, Lu YP, Lou YR, Kawasumi M, Nghiem P
Front Oncol 2013;3:144 [Review]
PMID: 23785666
ATR-Chk1 pathway inhibition promotes apoptosis after UV treatment in primary human keratinocytes: potential basis for the UV protective effects of caffeine
Heffernan TP, Kawasumi M, Blasina A, Anderes K, Conney AH, Nghiem P
J Invest Dermatol 2009;129(7):1805-1815
PMID: 19242509
Protection from photodamage by topical application of caffeine after ultraviolet irradiation
Koo SW, Hirakawa S, Fujii S, Kawasumi M, Nghiem P
Br J Dermatol 2007;156(5):957-964
PMID: 17388926
Book Chapter
Kawasumi M, Nghiem P. Chapter 112 “Ultraviolet Radiation Carcinogenesis”. In Fitzpatrick’s Dermatology in General Medicine, Eighth Edition (eds. Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, Wolff K), pp. 1251-1260. McGraw-Hill, USA, 2012.
Media Appearances
September 29, 2017
“Can Your Coffee Habit Help Protect You From Skin Cancer?” in UW Medicine Right as Rain.
July 2, 2015
“Skin cancer: 9 things to know to lower your risk” in Fred Hutch News.
August 15, 2011
“UW Medicine study finds caffeine guards against certain ultraviolet-induced skin cancers at molecular level” in UW News.
ATR Inhibitors
How can we target the ATR kinase signaling pathway to modulate DNA damage responses and treat cancer?
Selective killing of cancer cells by ATR inhibition
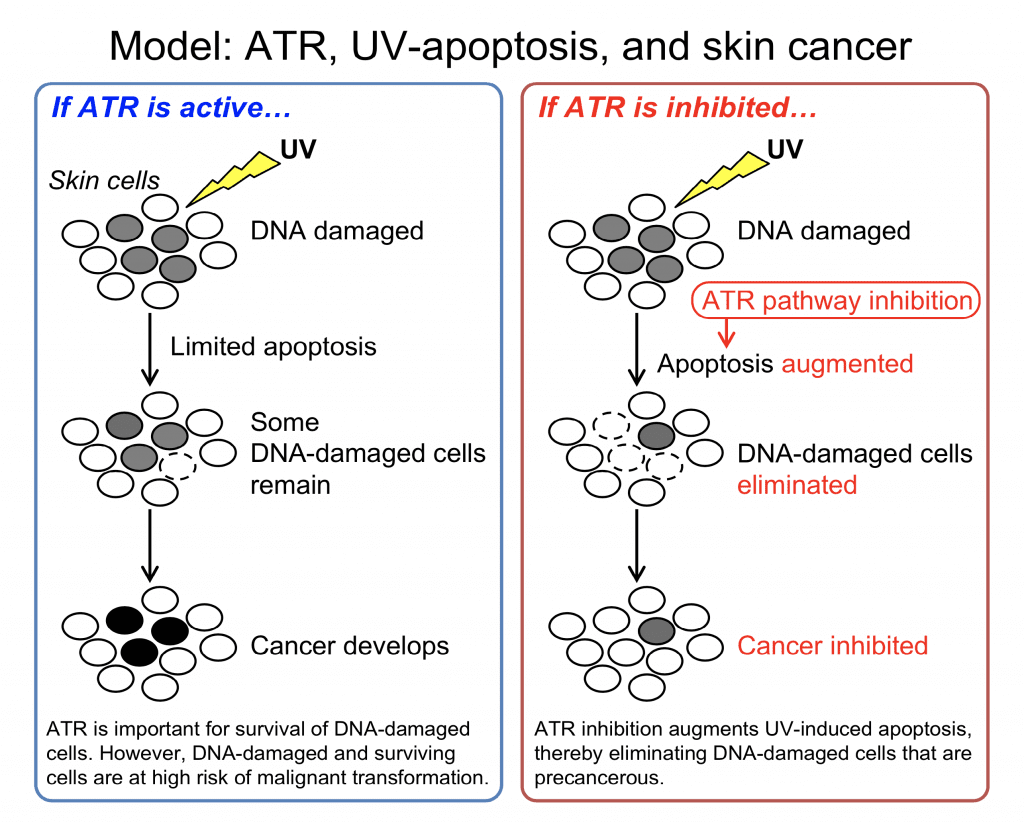
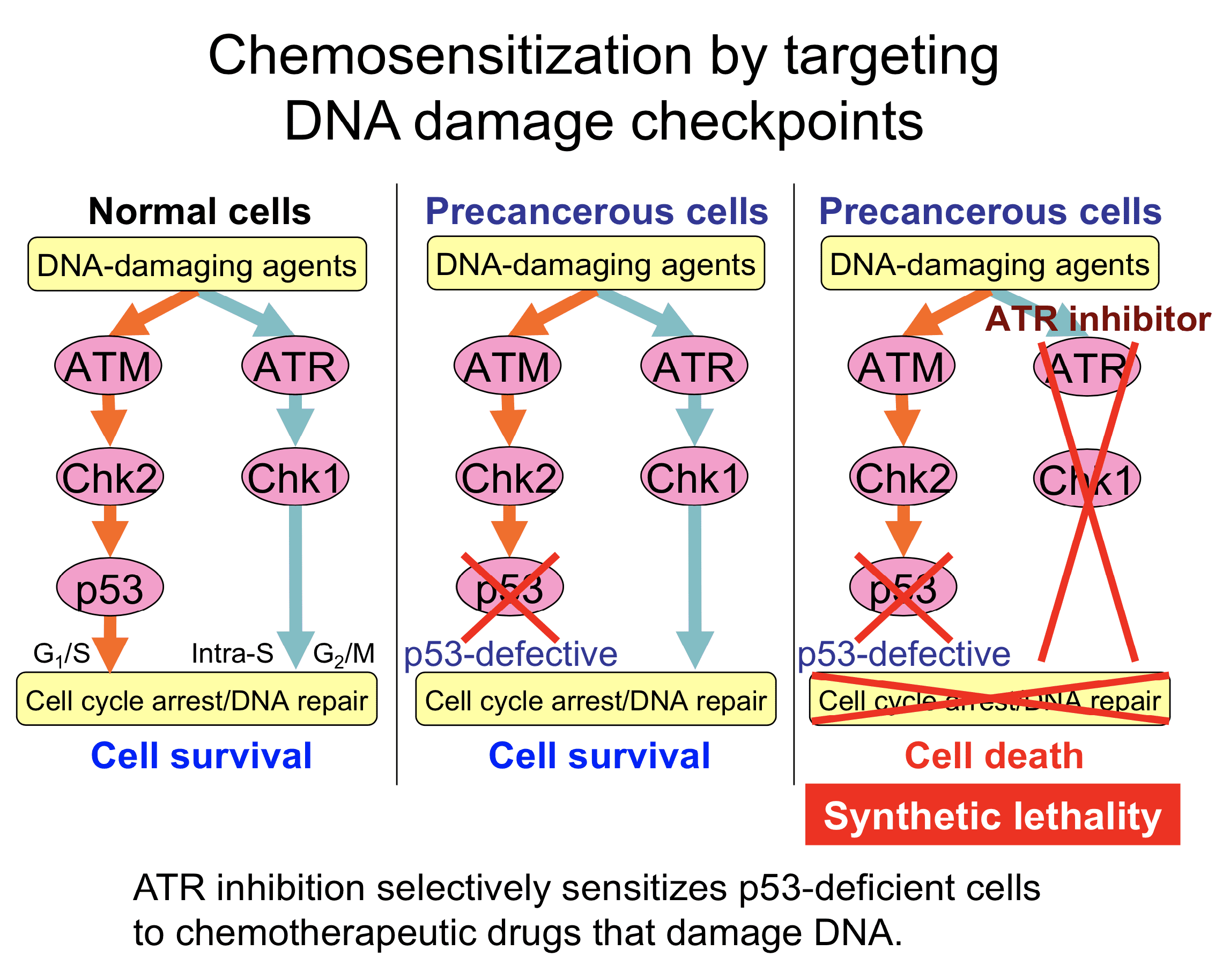
ATR is a pivotal kinase that senses UV damage and activates DNA damage checkpoint to stop the cell cycle. This allows time for DNA repair and promotes cell survival. ATM is a kinase critical for sensing DNA double-strand breaks. Many chemotherapeutic drugs are DNA-damaging agents and activate both ATM-Chk2-p53 and ATR-Chk1 pathways.
Targeting the ATR pathway is an appealing strategy to selectively kill cancer cells while sparing normal cells. Many cancer cells are p53-defective, thereby relying on the ATR pathway. When p53-deficient cells are treated with ATR inhibitors in the presence of DNA-damaging agents, neither ATM nor ATR pathway can stop the cell cycle, leading to cell death.
Small-molecule screening for novel ATR pathway inhibitors
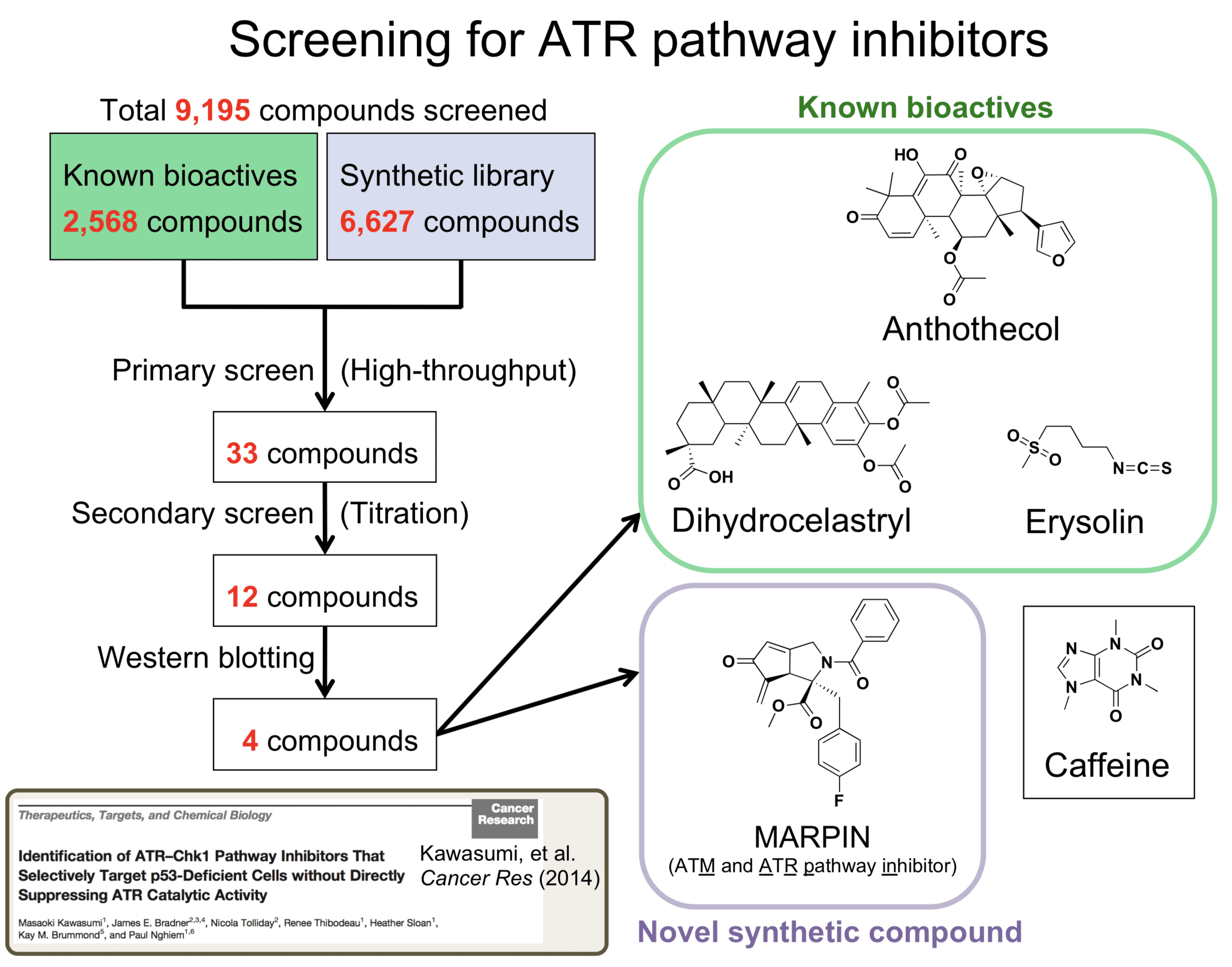
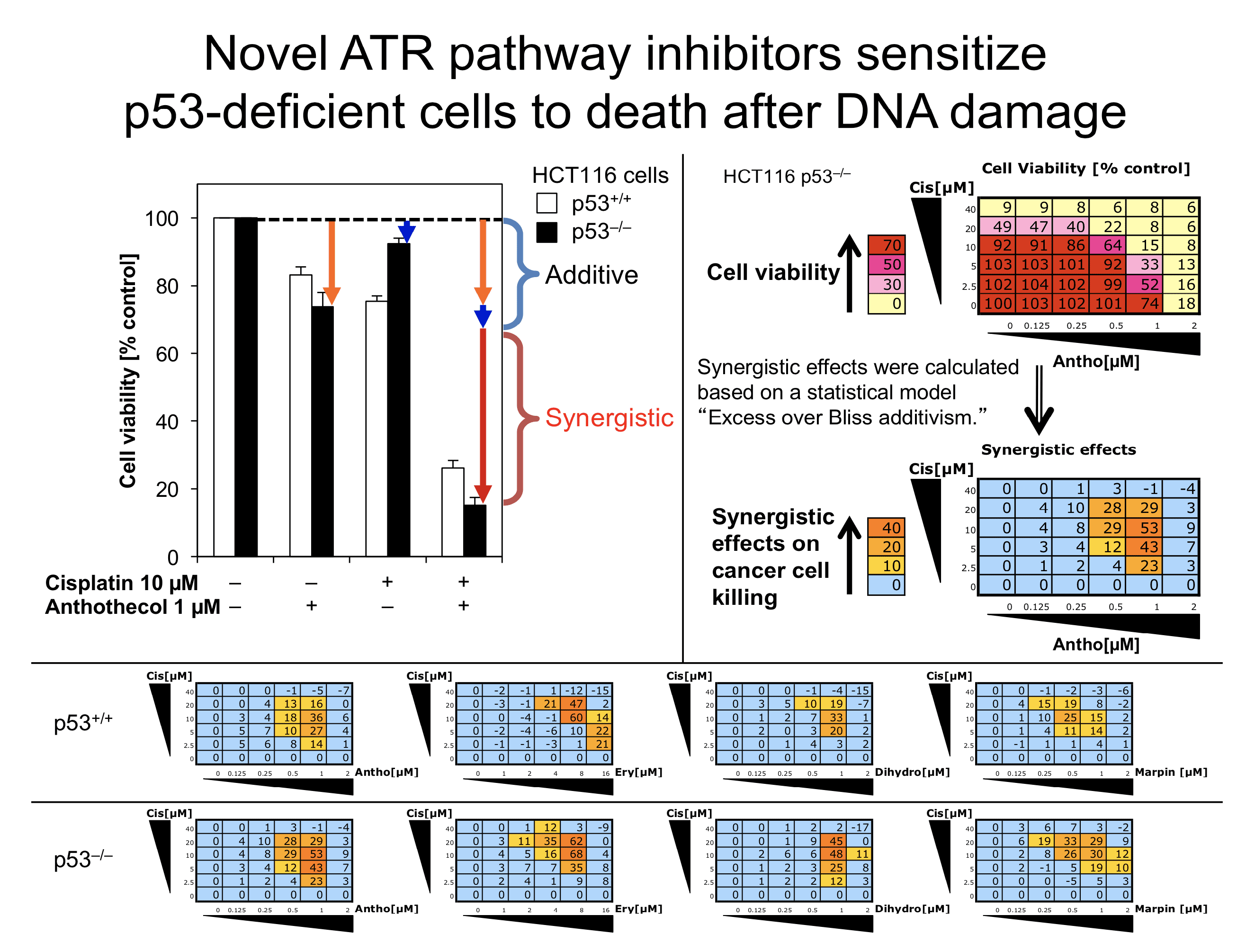
To discover druggable targets in the ATR pathway and novel classes of the ATR pathway inhibitors, we performed a phenotype-based screen of 9,195 small-molecule compounds and identified four compounds that inhibit the ATR pathway. These novel ATR pathway inhibitors sensitize p53-deficient cells to DNA-damaging agents (cisplatin and camptothecin) not only in vitro but also in an in vivo xenograft model.
Intriguingly, these ATR pathway inhibitors did not suppress ATR kinase activity in vitro, suggesting that these small-molecule compounds are mechanistically distinct from typical ATR kinase catalytic inhibitors. This study highlights the complexity of the ATR pathway, and these novel ATR pathway inhibitors can be used to further elucidate druggable mechanisms in order to improve cancer therapy. Currently, we focus on identifying a cellular target of MARPIN (ATM and ATR pathway inhibitor) that is readily derivatizable.
References
Identification of ATR-Chk1 pathway inhibitors that selectively target p53-deficient cells without directly suppressing ATR catalytic activity
Kawasumi M, Bradner JE, Tolliday N, Thibodeau R, Sloan H, Brummond KM, Nghiem P
Cancer Res 2014;74(24):7534-7545
PMID: 25336189
Chemical methodology as a source of small-molecule checkpoint inhibitors and heat shock protein 70 (Hsp70) modulators
Huryn DM, Brodsky JL, Brummond KM, Chambers PG, Eyer B, Ireland AW, Kawasumi M, Laporte MG, Lloyd K, Manteau B, Nghiem P, Quade B, Seguin SP, Wipf P
Proc Natl Acad Sci U S A 2011;108(17):6757-6762
PMID: 21502524
Chemical genetics: elucidating biological systems with small-molecule compounds
Kawasumi M, Nghiem P
J Invest Dermatol 2007;127(7):1577-1584 [Review]
PMID: 17568801
Epigenetics
How can we regulate gene expression to inhibit skin cancer?
Epigenome editing to inhibit skin cancer
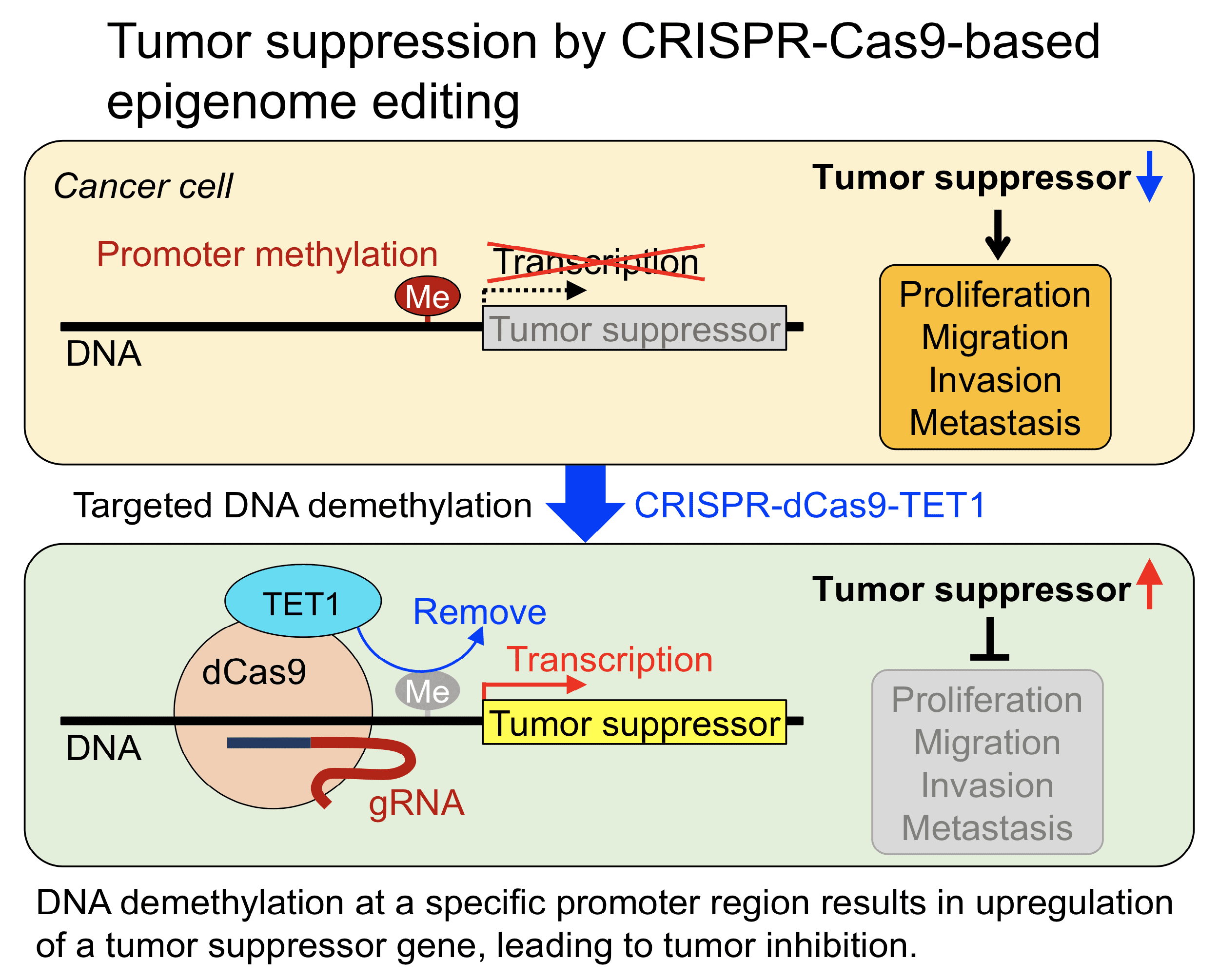
The p16INK4A and p14ARF tumor suppressor genes function as cell cycle regulators and play an important role in tumor growth and metastasis. These genes are frequently inactivated in skin cancers, mainly due to promoter methylation of p16INK4A and p14ARF rather than gene deletion or loss-of-function mutations. This DNA methylation is of therapeutic interest because epigenetic changes have the potential to be reversed to restore gene expression. We aim to develop novel epigenome editing tools to induce DNA demethylation at specific genomic loci in order to upregulate tumor suppressor genes and inhibit skin cancer. We will modify the CRISPR-Cas9 system to recruit DNA-modifying enzymes (e.g., TET1 for DNA demethylation) to specific genomic loci. We will determine cancer-inhibiting effects of DNA demethylation specifically at the p16INK4A and/or p14ARF promoter on proliferation, migration, invasion, and metastasis of skin cancer.
References
CRISPR-Cas9 epigenome editing to induce DNA demethylation at p14ARF promoter and inhibit skin cancer
Lee JW, Rokunohe D, Kawasumi M
J Investig Med 2018;66:109 [Meeting Abstract]
Ultraviolet (UV) irradiation
UV-induced DNA lesions
Cyclobutane pyrimidine dimers (CPD)
6-4 photoproducts (6-4PP)
DNA damage response (DDR)
DNA damage checkpoint
Cell cycle checkpoint
Replication checkpoint
ATR kinase
Caffeine
Small-molecule inhibitors of the ATR-Chk1 pathway
Chemical genetics
High-content screening
Chromatin recruitment of the ATR activation complex
Chemosensitization
Chemoprevention
Skin cancer prevention by caffeine
Nonmelanoma skin cancer
Melanoma
Xeroderma pigmentosum
DNA replication blockage at UV lesions
Error-prone translesion synthesis
UV mutagenesis
UV carcinogenesis
Next-generation sequencing
Duplex Sequencing
Epigenetics
The p16INK4A and p14ARF tumor suppressor genes
Promoter hypermethylation
CRISPR-Cas9
Epigenome editing
Targeted DNA demethylation
Systemic lupus erythematosus (SLE)