Highlights Merkel cell carcinoma (MCC) is an aggressive cutaneous malignancy with a recurrence rate of…
Highlights
- A high-content imaging screen was performed for discovery of ATR pathway inhibitors.
- Novel ATR pathway inhibitors do not directly suppress ATR catalytic activity.
- Novel ATR pathway inhibitors sensitize p53-deficient cells to DNA-damaging agents in vitro.
- A novel ATR pathway inhibitor synergizes with cisplatin to suppress p53-deficient tumor growth in vivo.
Abstract
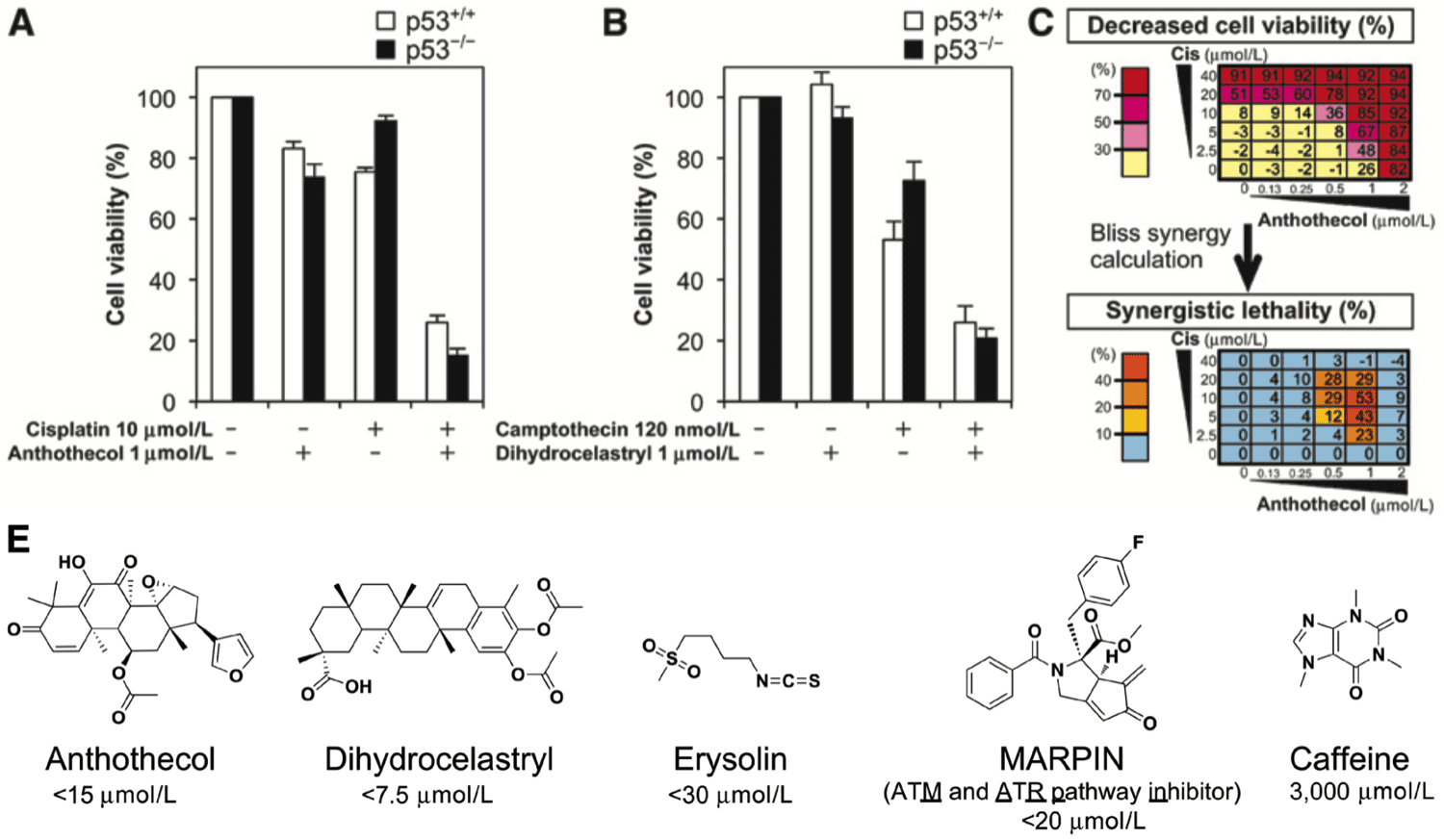
Resistance to DNA-damaging chemotherapy is a barrier to effective treatment that appears to be augmented by p53 functional deficiency in many cancers. In p53-deficient cells in which the G1-S checkpoint is compromised, cell viability after DNA damage relies upon intact intra-S and G2-M checkpoints mediated by the ATR (ataxia telangiectasia and Rad3 related) and Chk1 kinases. Thus, a logical rationale to sensitize p53-deficient cancers to DNA-damaging chemotherapy is through the use of ATP-competitive inhibitors of ATR or Chk1. To discover small molecules that may act on uncharacterized components of the ATR pathway, we performed a phenotype-based screen of 9,195 compounds for their ability to inhibit hydroxyurea-induced phosphorylation of Ser345 on Chk1, known to be a critical ATR substrate. This effort led to the identification of four small-molecule compounds, three of which were derived from known bioactive library (anthothecol, dihydrocelastryl, and erysolin) and one of which was a novel synthetic compound termed MARPIN. These compounds all inhibited ATR-selective phosphorylation and sensitized p53-deficient cancer cells to DNA-damaging agents in vitro and in vivo. Notably, these compounds did not inhibit ATR catalytic activity in vitro, unlike typical ATP-competitive inhibitors, but acted in a mechanistically distinct manner to disable ATR-Chk1 function. Our results highlight a set of novel molecular probes to further elucidate druggable mechanisms to improve cancer therapeutic responses produced by DNA-damaging drugs.